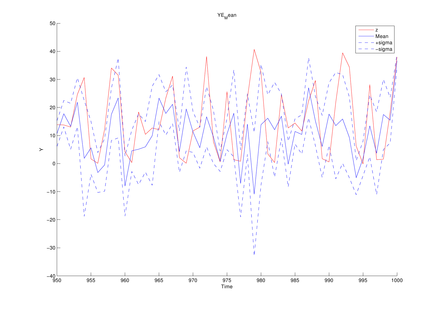
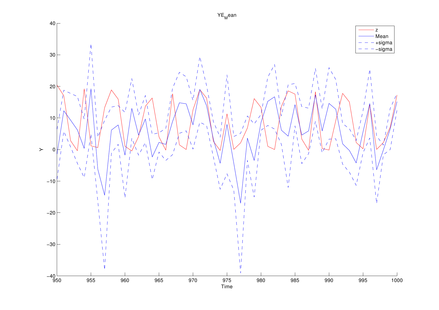
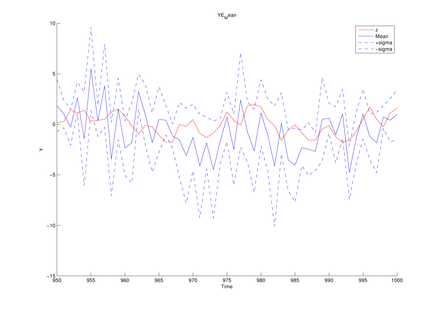
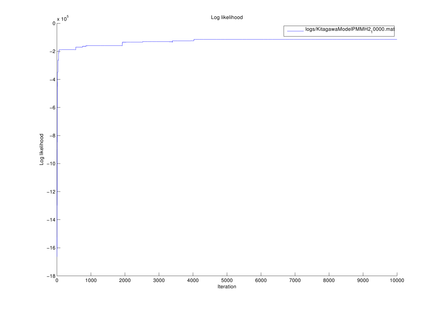
Here we introduce the example of model estimation using the MCDC tool, given observed sequences generated by the generalized Kitagawa model given by the following SSM:
- State transition function:
-
x1(t) = a1 * x1(t-1) + b1 * x1(t-1) / (1 + x1(t-1) ^ 2) + c1 * cos(1.2 * t) + sqrt(q1) * randn; x2(t) = a2 * x2(t-1) + b2 * x2(t-1) / (1 + x2(t-1) ^ 2) + c2 * cos(1.2 * t) + sqrt(q2) * randn;
- Observation function:
-
y1(t) = d1 * x1(t) ^ 2 + d2 * x2(t) ^ 2 + sqrt(r1) * randn; y2(t) = d3 * x1(t) ^ 2 + sqrt(r2) * randn; y3(t) = d4 * x2(t) + sqrt(r3) * randn;
where a1, a2, b1, b2, c1, c2, d1, d2, d3, and d4 are the
model static parameters. The parameters q1, q2, r1, r2, and r3
control the magnitude of state transition/observation noise.
| Parameter name |
Description |
Value |
|---|---|---|
| T | length of data sequence | 1,000 |
| (a1, b1, c1) | model parameters for state variable x1 | (0.5, 28, 8) |
| (a2, b2, c2) | model parameters for state variable x2 | (0.6, 30, 10) |
| (d1, d2, d3, d4) | model parameters for observation variable y |
(0.05, 0.06, 0.07, 0.08) |
| (q1, q2) | magnitude of noise added to state transition
function |
(1e-1, 1e-1) |
| (r1, r2, r3) | magnitude of noise added to observation function |
(1e-1, 1e-1, 1e-1) |