- State transition function:
-
x1(t) = (Sigma * x2(t-1) * dt + x1(t-1) * (1 - Sigma * dt)) + sqrt(q1) * randn; x2(t) = (x1(t-1) * (Rho - y1(t-1))) * dt + x2(t-1) * (1 - dt) + sqrt(q2) * randn;
- Observation function:
-
y1(t) = (x1(t-1) * x2(t-1) * dt) + y1(t-1) * (1 - Beta * dt) + sqrt(r1) * randn; y2(t) = Alpha0 * x1(t-1) .^ 2 + sqrt(r2) * randn; y3(t) = Alpha0 * x2(t-1) .^ 2 + sqrt(r3) * randn;
where Sigma, Rho, Beta, Alpha0 are the model static
parameters. The parameters q1, q2, r1, r2, and r3 control the
magnitude of state transition/observation noise.
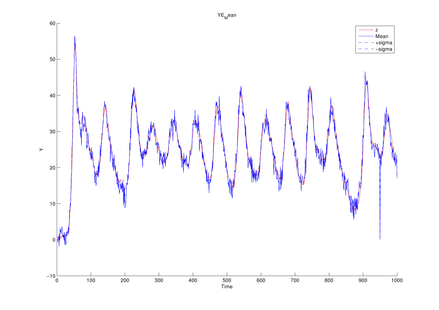
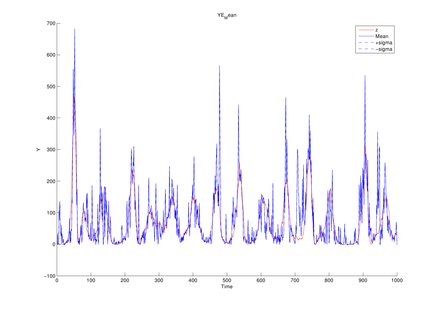
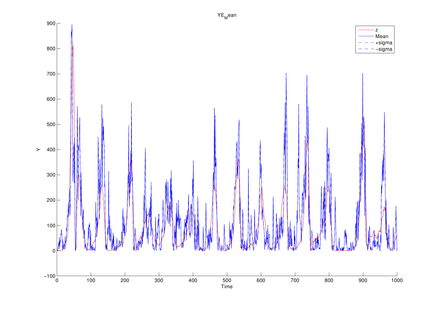
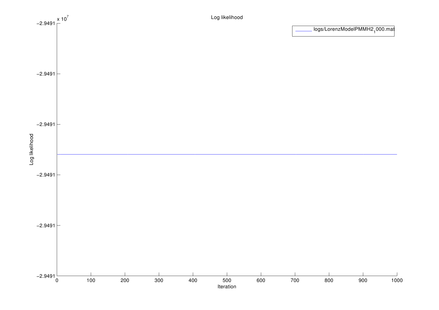
Parameter setting for Lorenz model
| Parameter name |
Description |
Value |
|---|---|---|
| T | length of data sequence |
1,000 |
| Sigma | model parameter | 10 |
| Rho | model parameter | 28 |
| Beta | model parameter | 8/3 |
| Alpha0 | model parameter | 1 |
| (q1, q2) | magnitude of noise added to state transition function | (1e-1, 1e-1) |
| (r1, r2, r3) | magnitude of noise added to observation function | (1e-1, 1e-1, 1e-1) |
| dt | time interval |
1e-2 |